InstaDeep is pleased to announce that a record five of its AI research papers have been accepted for presentation at the Thirty-sixth Annual Conference on Neural Information Processing Systems (NeurIPS 2022) with two papers in the main track and three workshop papers, including one authored in collaboration with BioNTech as part of our joint AI Innovation Lab.
NeurIPS 2022 is the 36th edition of this highly prestigious annual machine learning conference, with sessions and workshop tracks presenting the latest research in artificial intelligence and its applications in areas such as computer vision, computational biology, reinforcement learning, and more. InstaDeep’s continued participation in the conference over many years validates yet again our in-house research team’s capability to innovate and make a valuable contribution to the global AI research community.
Last but not least, NeurIPS 2022 also marks the first-ever accepted African female lead author, and first-ever accepted Tunisian lead author – both accolades for InstaDeep Research Engineer, Rihab Gorsane. Click here to read more about Rihab’s achievements and how InstaDeep is actively helping develop the AI Research ecosystem in Africa.
InstaDeep at NeurIPS 2022
The core focus at NeurIPS is peer-reviewed research which is presented and discussed in the general session, referred to as the main conference alongside invited talks by leaders in their field. Additionally, the conference supports a range of workshops where further research is presented.
Main Conference
This year, InstaDeep’s main conference contributions have been centred around research in intelligent multi-agent systems.
Towards a Standardised Performance Evaluation Protocol for Cooperative MARL
Rihab Gorsane1, Omayma Mahjoub2, Ruan de Kock1, Roland Dubb3, Siddarth Singh1, Arnu Pretorius1
1InstaDeep, 2National School of Computer Science, Tunisia, 3University of Cape Town, South Africa
Multi-agent reinforcement learning (MARL) has emerged as a useful approach to solving decentralised decision-making problems at scale. Research in the field has been growing steadily with many breakthrough algorithms proposed in recent years. Despite the fact that MARL is significantly less established and developed than single-agent RL, which may make the adoption of ethical evaluation approaches easier, the field is nevertheless afflicted by many of the same issues as the single-agent setting, such as implementation inconsistencies, irregular baselines, and a lack of statistical rigour in reported outcomes.
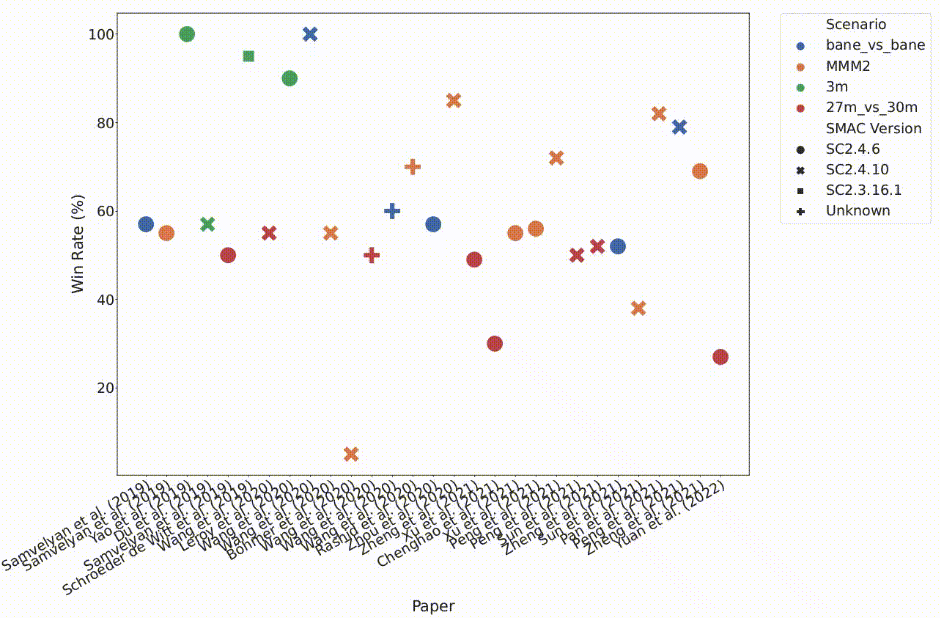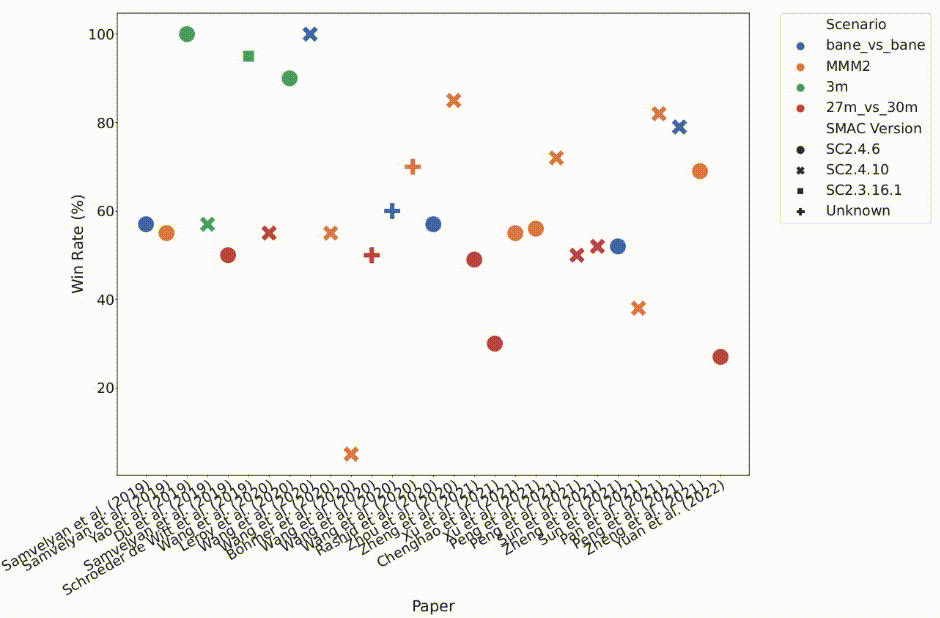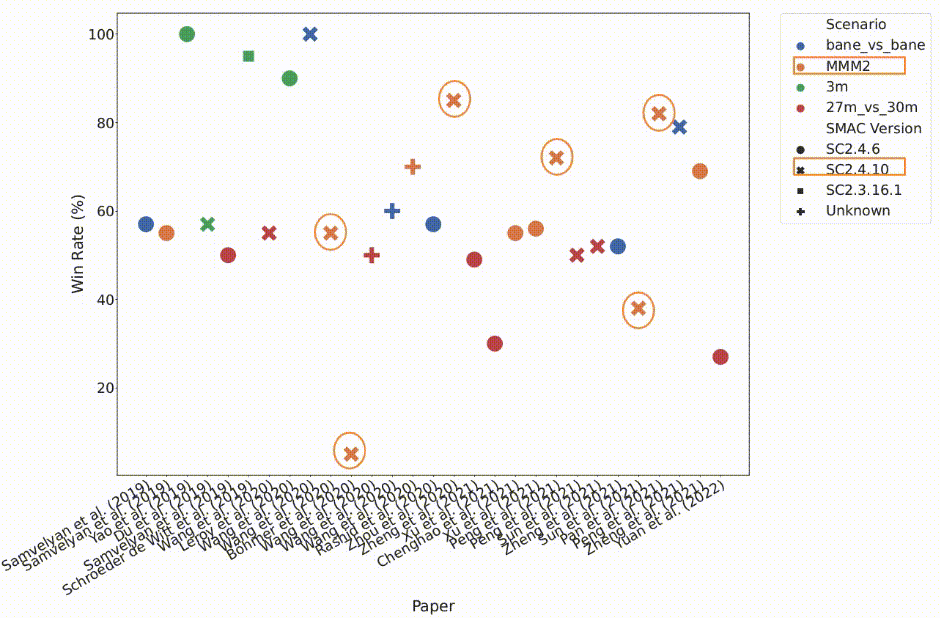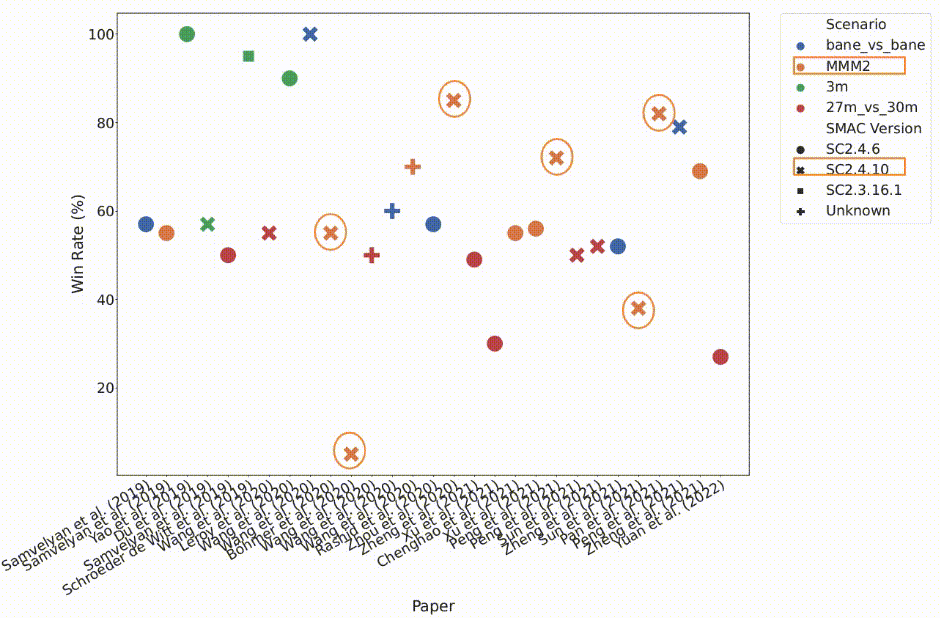
In this work, we take a closer look at the rapid development of MARL with a focus on evaluation methodologies employed across a large body of research. We bring to light worrying trends that put into question the true rate of progress (as shown in the above figure). We further consider these trends in a wider context and take inspiration from single-agent RL literature on similar issues with recommendations that remain applicable to MARL. Combining these recommendations, with novel insights from our analysis, we propose a standardised performance evaluation protocol for cooperative MARL. We argue that such a standard protocol, if widely adopted, would greatly improve the validity and credibility of future research, make replication and reproducibility easier, as well as improve the ability of the field to accurately gauge the rate of progress over time by being able to make sound comparisons across different works.
Links: Paper | Website | Open-Source Code | Poster session 4: Wed 30 Nov, 1400 – 1600 (PST), Hall J, #927
Universally Expressive Communication in Multi-Agent Reinforcement Learning
Matthew Morris1,2, Thomas D. Barret1, Arnu Pretorius1
1InstaDeep, 2University of Oxford, UK
Allowing agents to share information with each other through communication is crucial for solving complex tasks in multi-agent reinforcement learning (MARL). For example, trains need to communicate with one another to avoid entering the same section of track and colliding, robots moving supplies around may need to mutually agree on strategies for what/when to move, and agents searching through an environment may benefit from sharing their local findings with the other agents.
Different methods have emerged for achieving this, many of which can be described as Graph Neural Networks (GNNs). Our first contribution in this work is providing a unifying framework through which to view many of the most successful MARL communication methods, which we refer to as “Graph Decision Networks (GDNs)”. Since GDNs relate to standard GNNs, this allows us to leverage theoretical results from GNNs to show that there are types of communication protocols which GDNs are unable to learn. Furthermore, we found that GDNs are unable to perform “symmetry breaking”: i.e. they cannot solve problems which require similar agents that have identical observations to take different actions. For example, if there are a group of identical drones in a plain field, it is impossible for them to communicate using GDNs to split up and search the field.
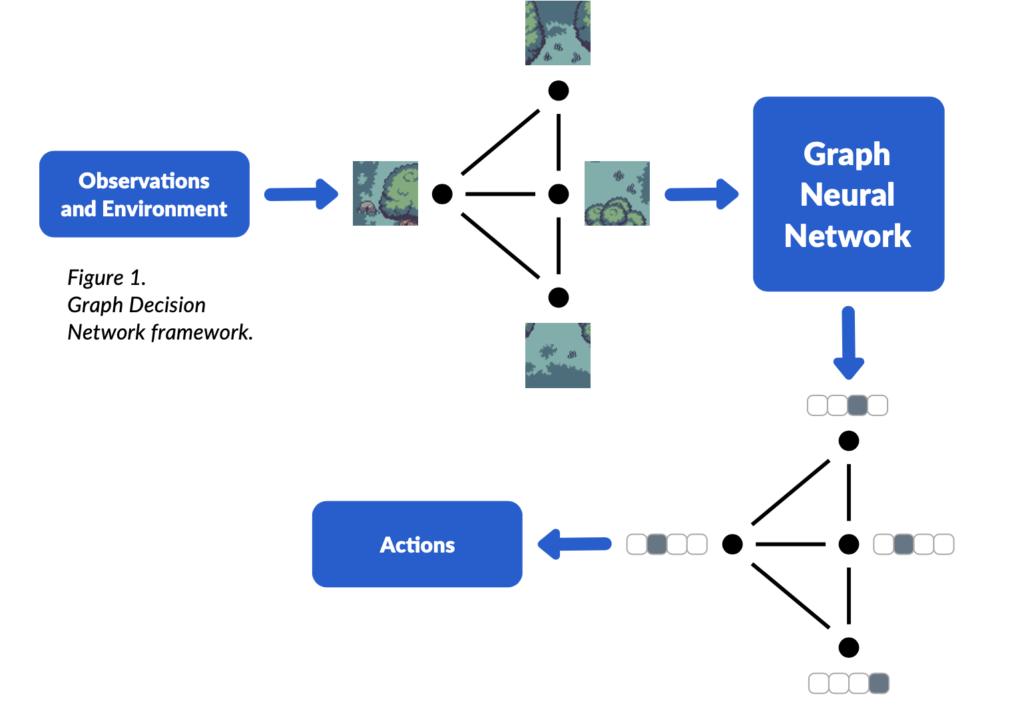
To solve these problems, we take two existing augmentations from GNNs and apply them to GDNs, appending 1) random noise and 2) unique IDs to each of the agent observations. We prove that a rich series of theoretical results hold for such augmented GDNs, despite the augmentations being very simple. We design two new multi-agent environments to test the expressivity of communication: 1) drones searching for a target in a field and 2) robots coordinating to move a box. We augment several different GDN models with random noise and unique IDs, doing an extensive evaluation, and find that, in general, the augmentations do not reduce performance. In our designed environments where complex communication is required, the augmented models greatly improve performance over the previous standard approaches.
Links: Paper | Code | Poster session 4: Wed 30 Nov, 1400 – 1600 (PST), Hall J, #118
NeurIPS Workshops
InstaDeep’s further contributions for 2022 are presented as part of specific workshops including participation in the AI for Science, Machine Learning in Structural Biology and Meta-Learning workshops.
So ManyFolds, So Little Time: Efficient Protein Structure Prediction with pLMs and MSAs
Thomas D. Barrett1, Amelia Villegas-Morcillo2, Louis Robinson1, Benoit Gaujac3, David Adméte1, Elia Saquand1, Karim Beguir1, Arthur Flajolet1
1InstaDeep, 2University of Granada, Spain, 3University College London, UK
In recent years, machine learning approaches for de novo protein structure prediction have made significant progress, culminating in AlphaFold which approaches experimental accuracies in certain settings and heralds the possibility of rapid in silico protein modelling and design. However, such applications can be challenging in practice due to the significant compute required for training and inference of such models, and their strong reliance on the evolutionary information contained in multiple sequence alignments (MSAs), which require expensive search of external sequences databases to generate and may not be available for certain targets of interest. Within the last year, an alternative to MSA-based models has emerged – leveraging information contained in pre-trained protein language models (pLMs) for downstream protein-folding tasks (e.g. ESMFold, OmegaFold). Whilst MSA-based models still provide SOTA accuracies, pLM-based approaches are closing the gap whilst not requiring external databases and showing signs of improved performance in low-, or no, MSA regimes.
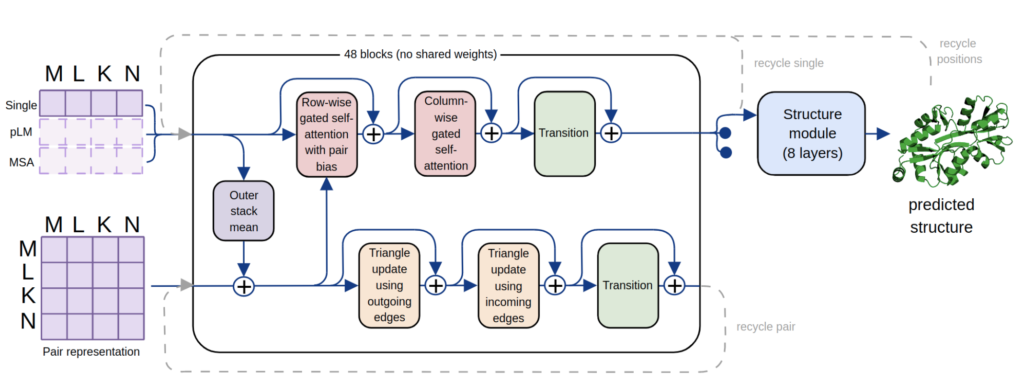
Our work presents a streamlined AlphaFold-like architecture and training pipeline, with which we trained equivalent models on either pLM embeddings or MSA statistics (MonoFold-pLM and MonoFold-MSA), and PolyFold – a single model capable of taking either, or both, pLM embeddings and MSAs as input. All models provide strong performance in only ~1 day of training time, with PolyFold working with all input modes as well as models trained uniquely those settings. In addition to the increased flexibility this provides, we also find that different targets are better predicted using different inference modalities, with none of the three options (pLM-only, MSA-only, pLM+MSA) either dominating or being uniformly inferior. Ultimately, this underlines the utility of our unified model and opens the door to further pushing the limits of performance with multiple complimentary sources of input information.
Links: Paper | AI for Science workshop | Machine Learning in Structural Biology | ManyFold Library
Peptide-MHC Structure Prediction With Mixed Residue and Atom Graph Neural Network
Antoine P. Delaunay1, Yunguan Fu1, Alberto Bégué1, Robert McHardy1, Bachir A. Djermani1, Michael Rooney2, Andrey Tovchigrechko2, Liviu Copoiu1, Marcin J. Skwark1, Nicolas Lopez Carranza1, Maren Lang2, Karim Beguir1 and Uğur Şahin2
1InstaDeep, 2BioNTech
In collaboration with BioNTech as part of the joint AI Innovation Lab, we have been working on structure prediction of the peptide-Major Histocompatibility Complex (pMHC). This protein complex plays an essential role in the T-cell-mediated immune response, making its understanding necessary to design future cancer therapies. This problem remains difficult to solve due to limited available experimental data.
Current physical models require significant computational time, and state-of-the-art deep learning models such as AlphaFold 2 need tuning of a large number of parameters. A more restricted deep learning approach can overcome both of these difficulties. For this, we used Graph Neural Networks (GNNs) with graphs at the residue and atomic levels to encode both information related to the peptide-MHC interaction and the local atomic geometry. Combining deep learning with biophysical knowledge is essential to design an accurate model as the biophysical coherence of the predicted structure must be considered.
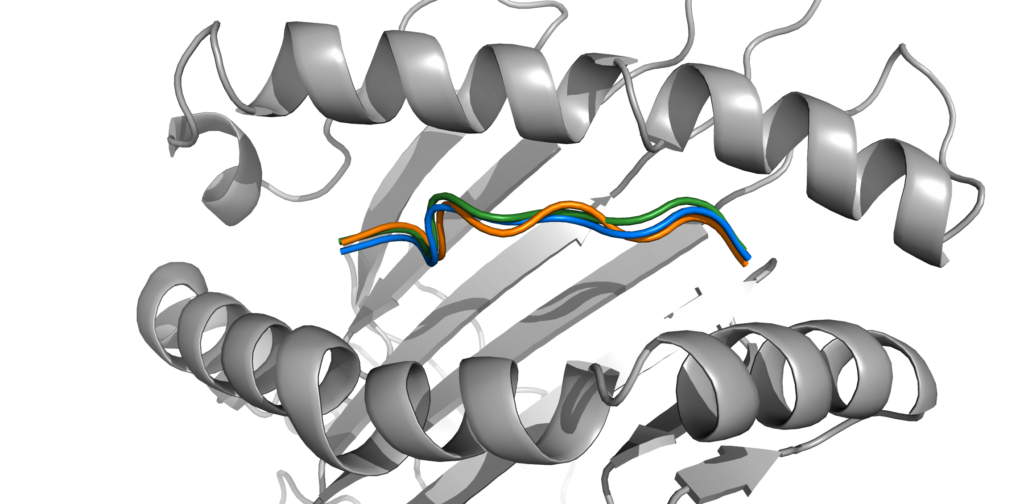
The results show that our model can reach comparable performances to other protein-structure prediction models, such as AlphaFold 2, with far fewer parameters (1.7M vs 93M). In particular, the GNN predictions have a similar coordinate accuracy but a higher biophysical consistency. The model remains restricted to class I MHC and 9-mer peptides, but future research will include extending it to increase the scope of pMHC complexes it can handle.
Links: Paper | Machine Learning in Structural Biology
Debiasing Meta-Gradient Reinforcement Learning by Learning the Outer Value Function
Clément Bonnet1, Laurence Midgley1, Alexandre Laterre1
1InstaDeep
Current state-of-the-art model-free reinforcement learning methods self-tune their hyper-parameters online. This means they automatically adapt the hyper-parameters used by their objective during training to improve learning. This meta-learning class of algorithms relies on an inner loss that uses meta-parameters, and an outer loss that is used to compute a meta-gradient with respect to these meta-parameters.
In this paper, we highlight a bias in the meta-gradient of current self-tuning methods. More specifically, the outer loss estimate is biased as it necessitates the value of the policy discounted with the outer loss discount factor but uses the critic that models the value function discounted by the different meta-learned discount factor. This bias in the advantage of the outer loss directly causes a meta-gradient bias that may favor myopic policies.
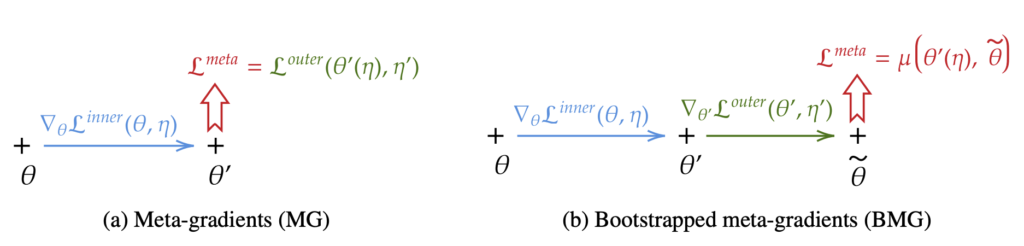
To help alleviate this issue, we propose to use the outer value function in the estimation of the outer loss that we learn by adding a second head to the critic network trained alongside the classic critic, yet using the outer loss discount factor. We apply our method to two different tasks and demonstrate that fixing the meta-gradient bias can significantly improve performance.
Links: Paper | Code | Meta-Learning workshop
Interested in our research work? Please consider joining us! Visit our careers page to see all the opportunities we currently have. We are hiring full-time and interns all over the year in most of our offices around the world! 🌎
