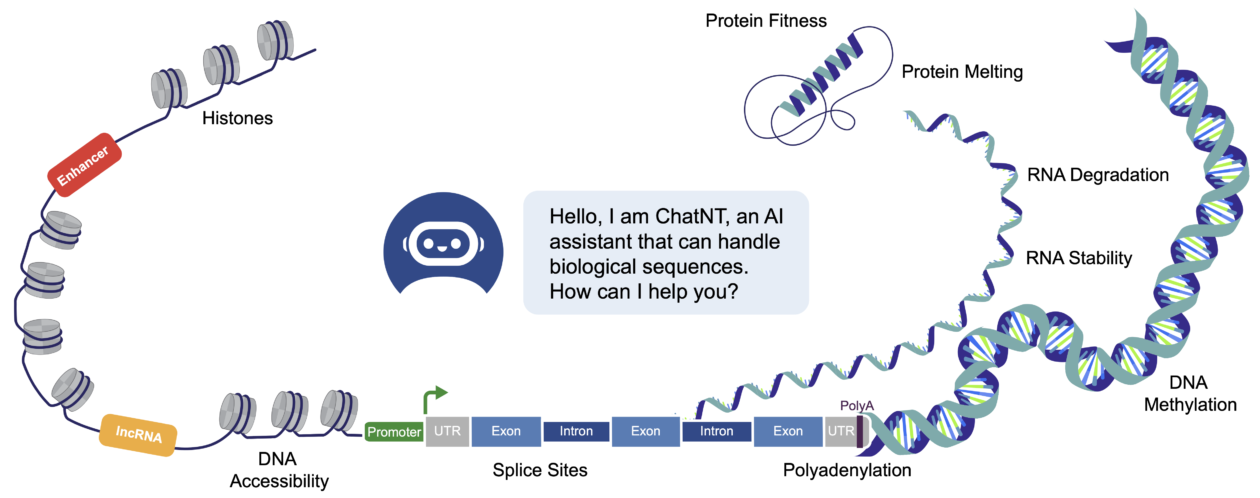
The human genome, containing the entirety of our genetic blueprint, holds the keys to understand the intricate workings of our cells and bodies. How our cells respond to signals, build proteins, and fight off diseases, is determined by instructions encoded within our DNA. While scientists have made tremendous progress in deciphering this code, especially since the complete mapping of the human genome, our understanding remains limited in many cases. The massive influx of data generated by modern sequencing techniques and assays presents both an amazing opportunity and a new challenge. This is where the power of Artificial Intelligence (AI) models comes into play, helping us process mountains of data to uncover hidden patterns within our genome.
Scientists have leveraged Deep Learning for the last ten years to build all sorts of predictive models for sequences of nucleotides, the base components of DNA. These models can predict important features such as (1) the location of regulatory elements, that can switch on and off the expression of genes, (2) the binding sites of proteins on the DNA and RNA, important in the regulation of different processes, (3) the accessibility and chromatin state of DNA regions in different cell types, a marker for their regulatory activity, or (4) the levels of expression of genes in different tissues (Eraslan et al. 2019; Yue et al. 2023). These models have already shown impressive improvements over previous statistical and machine learning methods and are now used in multiple important applications.
The Nucleotide Transformer: from supervised to self-supervised learning
Previous methods predominantly relied on supervised learning, which necessitates labeled data. Recently, the field of AI has been revolutionized by the emergence of self-supervised learning techniques, which allow for learning solely from unlabeled data. This development has facilitated the creation of advanced AI systems such as GPT-4, Gemini, and other similar systems (Achiam et al. 2023, Gemini team et al. 2023). The simultaneous decrease in genome sequencing costs, producing vast quantities of unlabeled genomic data, presents therefore an appealing opportunity to apply self-supervised learning to genomics. Several teams, including our own at InstaDeep, have adopted this strategy and developed genomic foundation models using self-supervised training. The process is simple: genomes from humans and various other species are cut into chunks, nucleotides are grouped into token-like representations, and these tokens are then used in common self-supervised learning systems to learn meaningful information from the data.
Motivated by the bi-directional nature of the information flowing through DNA, our models were trained using BERT, a technique in which some tokens are masked and the network is trained to reconstruct them from the surrounding context. As we relied on the Transformer architecture, probably the most popular and performing architecture in AI right now when it comes to sequence processing, we called our models Nucleotide Transformer (Dalla-Torre et al. 2023). We trained different model variants varying the genomes used for pre-training as well as the model sizes and the training duration, with our bigger model so far having 2.5 billion parameters. All our models were trained on large computing infrastructure, such as the Cambridge-1 cluster in the UK or a Google TPU-v4 superpod, for weeks. An important finding was that during pre-training our models acquired biological knowledge despite any supervision or labels. Notably, when investigating the activations and attention maps of our Nucleotide Transformer models, we observed that they learned to capture important patterns and sequence features such as detecting regulatory elements or differentiating between coding and non-coding regions. Thanks to the rich representation that these models acquire from DNA during pre-training, they can also be used as starting points to build new predictive models very quickly and at low cost through finetuning. This has made these models very popular as it allows researchers to try deep learning easily on their problems without having to think about different architectures or to spend the price of training a new model from scratch. The Nucleotide Transformer is, at the time we are writing this article, the most downloaded genomics AI model in the world, with up to 170k monthly downloads on Hugging Face, reflecting its popularity and accessibility for researchers new to deep learning.
Fine-tuning is a powerful technique, enabling models to adapt to new downstream tasks with minimal labeled data by leveraging knowledge gained from self-supervised pre-training. However, this approach can lead to a proliferation of models as the number of downstream tasks increases. This creates challenges in memory and data management, and hinders potential transfer learning opportunities between related tasks. In the quest to build generally capable biological research agents, achieving transfer learning is crucial – not only between unlabeled and labeled data but also across different labeled data sources. These sources include assays like ChIP-seq, ATAC-seq, STARR-seq and RNA-seq, conducted on various species and tissues. Since biological processes within a species are interconnected, and fundamental processes are conserved across species, transfer learning holds immense potential for maximizing knowledge gained from these varied datasets.
ChatNT: The first multimodal conversational agent for DNA, RNA and protein tasks
The natural language processing (NLP) field faced a similar challenge in 2017. The solution was to reframe diverse tasks as sets of questions and answers expressed in natural language, such as English. Sequence-to-sequence models were then trained to predict answers based on these questions. Introduced initially in the T5 project (Raffel et al. 2020), this technique saw extensive use in the GPT line of work (Brown et al. 2020), ultimately leading to advanced agents like ChatGPT (OpenAI, 2023). Inspired by this success, we have asked ourselves: “Could a similar technique be used to unify all downstream tasks of interest involving nucleotide sequences? And could natural language itself be also used here to formulate both questions and answers?”. The answer was a resounding “yes”!
To achieve this, we drew inspiration from recent techniques in creating multimodal models and developed the first multimodal DNA/language agent, called Chat Nucleotide Transformer (or ChatNT) (Richard et al. 2024). Techniques in multimodal language-vision models connect pre-trained vision encoders (like CLIP) (Radford et al. 2021) with large language models (like GPT), training the combined model end-to-end on “visual instructions” (Liu et al. 2024) – examples of questions and answers about images. In our case, we found out that we can do something similar, replacing the vision by a DNA encoder (the Nucleotide Transformer for instance) and replacing the image data by questions and answers relative to nucleotide sequences and biological processes. We used an instruction-finetuned Llama (Touvron, et al., 2023, Peng, et al., 2023) as a large language model and used a perceiver-resampler (Jaegle et al. 2021, Alayrac et al. 2022) as a projection layer model to transform the embeddings computed by the Nucleotide Transformer into a fixed set of embeddings in the natural language input space of the LLM. Keeping this modular architecture allows us to use constantly improving encoders and decoders in the future without changing the model architecture.
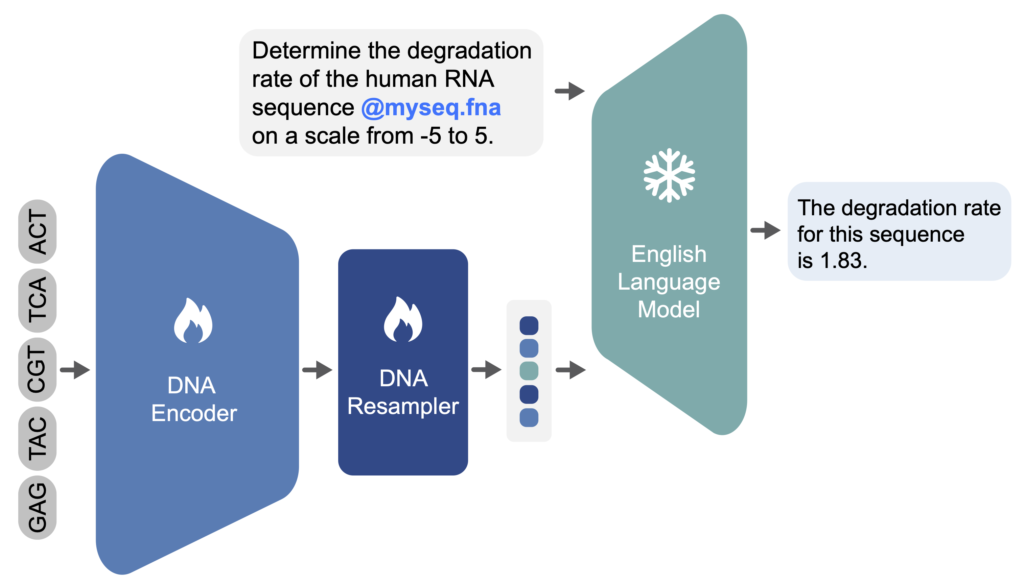
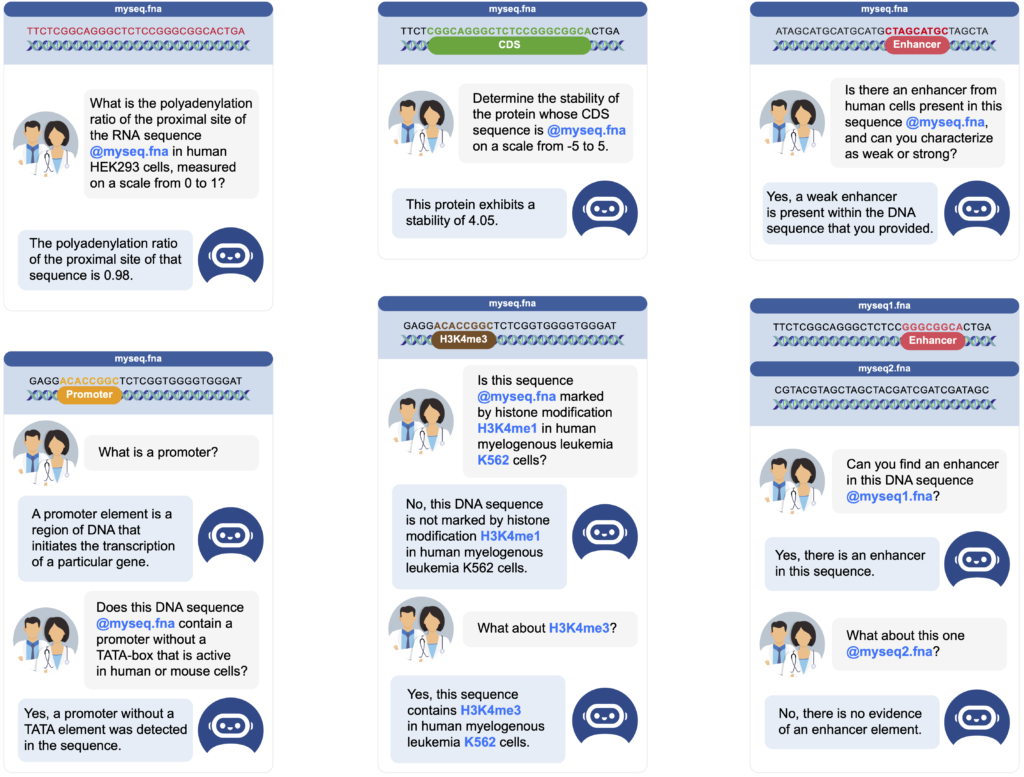
To train this system effectively, we needed to gather extensive data. The central dogma of biology establishes that DNA contains the blueprint for all cellular processes. This means that problems in genomics, RNA function, and protein structure can ultimately be addressed from their corresponding DNA sequences. Inspired by this principle, we expanded our model’s scope to evaluate its ability to tackle tasks related to proteins and RNA in addition to its core genomic functions. To achieve this, we carefully curated datasets commonly used to build deep learning models for DNA, RNA, and protein analysis. For each dataset, we generated a variety of questions and answers, yielding an average of 1,000 question/answer pairs per task. In total, we compiled over 40 tasks covering diverse biological processes and modalities – all fundamentally derived from DNA sequences. All sequences and their annotations originated from recently published assays, while our team newly curated all questions and answers. This resulted in a dataset comprising a total of 605 million DNA tokens, i.e. 3.6 billion base pairs, and 273 million English tokens. After training on the combined dataset, ChatNT achieved performance comparable to multiple specialized models on most tasks while being able to solve all tasks at once, in English, and to generalize to unseen questions.
ChatNT represents an innovative approach in biology, aiming to act as a universal translator for the language of our cells. Instead of relying on a collection of highly specialized, task-specific models, ChatNT unifies AI expertise in biology with an understanding of plain English. This means researchers can provide ChatNT with DNA sequences and directly ask questions like, “Will this DNA sequence likely make a protein glow green?” or “In which tissues might this gene be active?”. ChatNT then processes both the raw DNA data and the English instructions to generate accurate and informative responses. Beyond its current capabilities, ChatNT’s true potential lies in its adaptability. As it encounters new tasks and information, ChatNT learns and expands, opening up exciting possibilities for unifying biological research.
The Future of Genomics
The development of ChatNT signals a potential shift towards the creation of a truly universal multimodal genomics AI system – a comprehensive and adaptable tool empowering scientists and researchers. This could lead to streamlined workflows, accelerated discovery in the understanding of diseases and genetic risks, and ultimately open the door for the development of highly personalized medical treatments tailored to an individual’s unique DNA.
Download the full paper
References
Eraslan et al. “Deep learning: new computational modelling techniques for genomics.” Nature Reviews Genetics 20, 389–403 (2019).
Yue et al. “Deep Learning for Genomics: From Early Neural Nets to Modern Large Language Models.” Int. J. Mol. Sci. 2023, 24(21), 15858.
Dalla-Torre et al. “The nucleotide transformer: Building and evaluating robust foundation models for human genomics.” bioRxiv (2023): 2023-01.
Raffel et al. “Exploring the limits of transfer learning with a unified text-to-text transformer.” Journal of machine learning research 21.140 (2020): 1-67.
Touvron et al. “Llama: Open and efficient foundation language models.” arXiv preprint arXiv:2302.13971 (2023).
Peng et al. “Instruction tuning with gpt-4.” arXiv preprint arXiv:2304.03277 (2023).
Jaegle et al. “Perceiver: General perception with iterative attention.” International conference on machine learning. PMLR, 2021.
Alayrac et al. “Flamingo: a visual language model for few-shot learning.” Advances in neural information processing systems 35 (2022): 23716-23736.
Liu et al. “Visual instruction tuning.” Advances in neural information processing systems 36 (2024).
Radford et al. “Learning transferable visual models from natural language supervision.” International conference on machine learning. PMLR, 2021.
OpenAI. “GPT-4 technical report.” arXiv preprint arXiv:2303.08774 (2023).
Team Gemini et al. “Gemini: a family of highly capable multimodal models.” arXiv preprint arXiv:2312.11805 (2023).
Brown et al. “Language models are few-shot learners.” Advances in neural information processing systems 33 (2020): 1877-1901.