AI is revolutionising proteomics and has the potential to unlock new frontiers in targeted healthcare and biomedical research. At the heart of this is peptide sequencing, an essential process for understanding proteins and their role in biological systems.
Why Peptides?
Peptides are the building blocks of proteins. Accurately sequencing them drives advancements in drug discovery, biomarker identification, immunology, and disease diagnostics.
However, traditional sequencing methods rely on matching mass spectrometry data to protein databases, which limits their ability to identify unknown or novel proteins.
De novo peptide sequencing reconstructs peptide sequences without relying on a reference database, making it essential for identifying previously uncharacterised peptides. By directly interpreting mass spectrometry data, it uncovers novel proteins and modifications that traditional methods would miss.
Yet, existing de novo models struggle with accuracy and efficiency, often due to noisy mass spectrometry data and complex peptide modifications.
Our latest AI developments, InstaNovo and InstaNovo+, introduce an alternative approach to de novo sequencing. Featured in Nature Machine Intelligence, these models overcome the limitations of existing methods—enhancing accuracy, expanding discovery, and redefining what’s possible in peptide sequencing.
What Are InstaNovo and InstaNovo+?
InstaNovo is a transformer-based model designed for de novo peptide sequencing. Developed in collaboration between InstaDeep and the Department of Biotechnology and Biomedicine at the Technical University of Denmark (DTU), it translates fragment ion peaks from mass spectrometry data into peptide sequences with unprecedented precision.
Unlike traditional methods that rely on pre-existing databases, InstaNovo identifies peptides that have never been documented before—expanding the landscape of proteomic discovery.
A key innovation of the InstaNovo models is InstaNovo+, a diffusion-based iterative refinement model that enhances sequence accuracy by mimicking how researchers manually refine peptide predictions. InstaNovo+ begins with an initial sequence—either derived from InstaNovo or generated at random—and improves it, step by step.
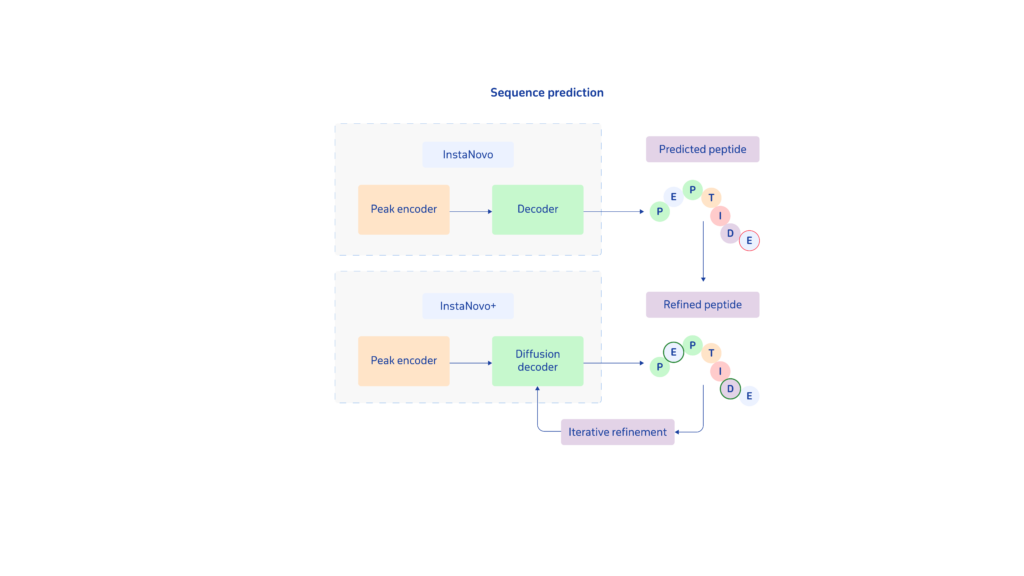
When paired with InstaNovo, InstaNovo+ significantly reduces false discovery rates (FDR) and improves sequence accuracy, not just by refining predictions, but by exploring a broader range of potential peptide sequences.
Unlike autoregressive models such as InstaNovo and Casanovo, which predict peptide sequences one amino acid at a time, InstaNovo+ processes entire sequences holistically, enabling greater accuracy and higher detection rates.
Together, InstaNovo and InstaNovo+ enhance de novo peptide sequencing, striking a balance between precision and exploration to accelerate biological discovery.
How Do They Work?
Peptide sequencing begins with mass spectrometry, where a sample is ionised and fragmented into smaller peptides. The mass-to-charge ratio of these fragments is then recorded, generating a mass spectrum that serves as the basis for sequence identification.
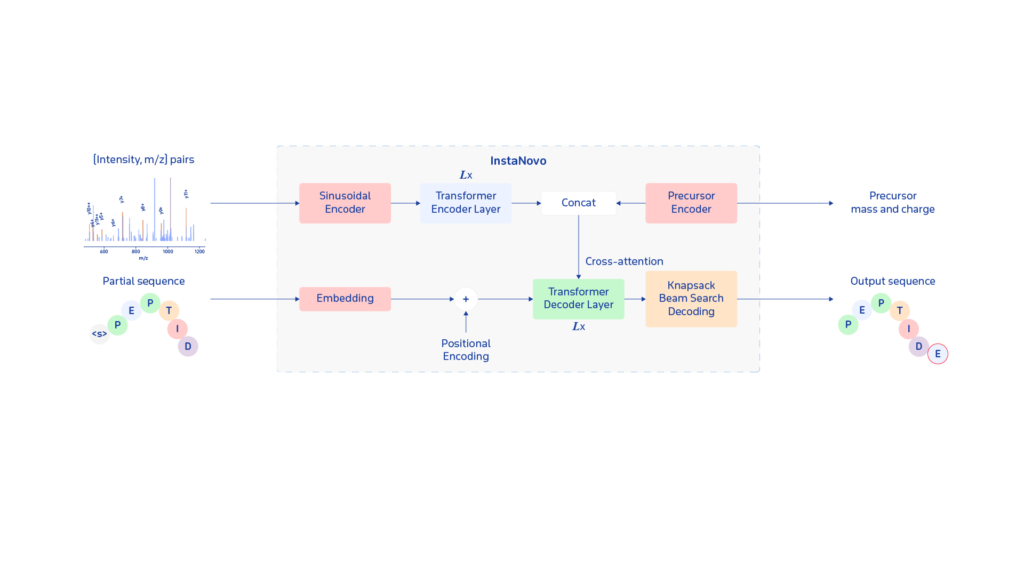
InstaNovo interprets mass spectra by mapping fragment ion peaks to peptide sequences, much like speech recognition converts audio into text. Instead of transcribing sound, InstaNovo predicts the next amino acid in a peptide sequence from mass spectrometry data, even without a reference database. InstaNovo+ then refines these predictions, ensuring alignment with real-world proteomic data.
To improve accuracy and reduce false positives, InstaNovo incorporates Knapsack Beam Search decoding into its architecture. This approach prioritises peptide sequences that fit the precursor mass constraints, reducing errors and increasing confidence in identifications.
By combining mass spectrometry with deep learning, InstaNovo and InstaNovo+ provide a scalable, AI-driven approach for de novo peptide sequencing, enabling more reliable and comprehensive proteomic discoveries.
Why Do We Need InstaNovo?
By eliminating dependency on protein databases and improving accuracy through iterative refinement, InstaNovo and InstaNovo+ uncover previously inaccessible proteomic landscapes, with the potential to drive discoveries across multiple scientific domains.
Even well-studied samples like HeLa cells contain novel proteins that traditional methods fail to detect. In testing, InstaNovo identified 1,338 previously undetected protein fragments, expanding the database and deepening our understanding of human proteomics.
In addition to novel peptides, InstaNovo enables high-precision sequencing of biomolecules such as nanobodies—small but powerful antibody fragments with the promise for targeted drug development. By accurately characterising their sequences, InstaNovo optimises nanobody binding to specific disease targets, potentially paving the way for treating infectious diseases, autoimmune disorders, and cancer.
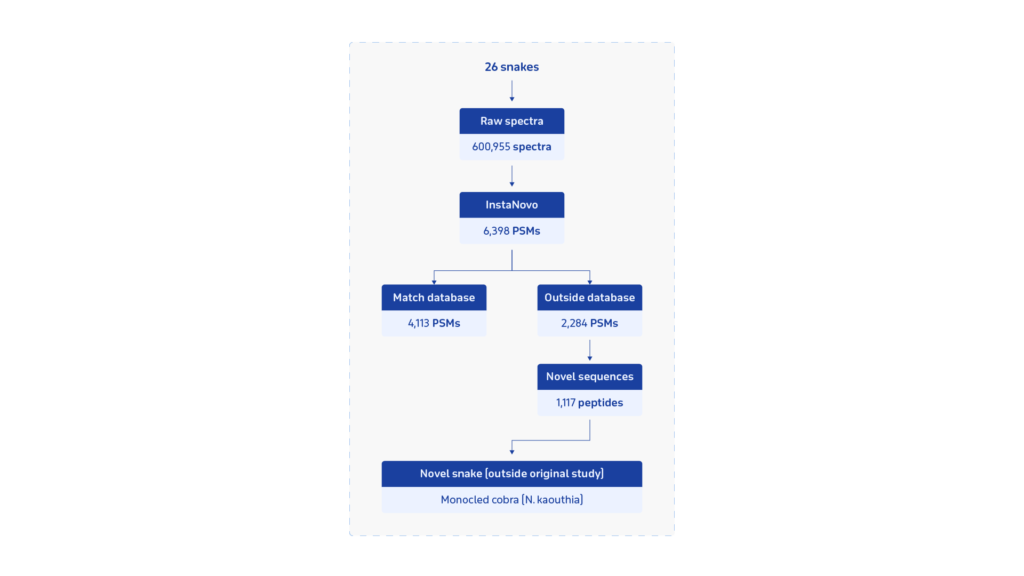
InstaNovo has also demonstrated success in identifying organisms in complex samples, such as undetected bacteria in wound fluid exudates—crucial for understanding infections and prescribing the right antibiotics. This capability extends to more complex biological mixtures, such as snake venoms. In testing, InstaNovo increased peptide spectrum matches (PSM) in snake venom samples by 20% and even detected venoms from species outside the original experiment scope (Figure 3).
Beyond sequencing applications, we believe InstaNovo has the capacity to enhance diagnostic accuracy through its analysis of the immunopeptidome—the collection of peptides displayed by Major Histocompatibility Complex (MHC) molecules on cell surfaces, essential for immune surveillance. Understanding these peptides informs the development of improved diagnostic tools and immunotherapies. InstaNovo+ advances this by enhancing the detection of MHC-bound peptides, aiding cancer immunotherapy and vaccine design. By identifying 12,965 novel PSMs, InstaNovo+ significantly expands the known immunopeptidome, offering real-time insights into diseases at a cellular level.
What’s Next?
InstaNovo has the potential to be a powerful catalyst for discovery. While not a replacement for rigorous scientific research, its enhancements could support progress across medicine, environmental science, and beyond.
We plan to expand InstaNovo’s training datasets, improving its peptide prediction across diverse species and environments. Additionally, future iterations may support a broader range of post-translational modifications (PTMs), deepening its role in proteomics by detecting both natural and induced modifications.
In addition, the development of user-friendly platforms should make InstaNovo more accessible to researchers, helping to advance protein science. By expanding its reach and refining its capabilities, InstaNovo could become an essential tool for advancing proteomics and accelerating biological discovery.
Disclaimer: all claims made are substantiated by our research paper: InstaNovo enables diffusion-powered de novo peptide sequencing in large scale proteomics experiments unless explicitly cited otherwise.
Go beyond the database with InstaNovo, today !
🚀 Experience AI-powered sequencing at your fingertips, with these powerful algorithms—Try InstaNovo now
Media enquiries please email: communications@instadeep.com
