Digital Biology
Our Digital Biology team leverages advanced machine learning and simulation techniques to revolutionise drug discovery at its core. Collaborating with domain experts, to transform intricate biological data into actionable insights, the team drives breakthroughs in genomics, proteomics, quantum chemistry, and beyond
Decision-Making
Our Decision-Making team pioneers reinforcement learning methods. By building AI systems that drive real-world impact—from chip design to resource management and scientific discovery—the team ensures the next generation of AI agents excels at dynamic decision-making.
ML Systems
Our Machine Learning (ML) systems team transform AI ambitions into practical, adaptable advancements. Driving breakthroughs from foundation models in biology to cutting-edge scientific computing, they tackle novel system challenges in AI infrastructure, enabling efficient large-scale ML algorithms.
Machine Learning
Our Fundamental Machine Learning (ML) team push the boundaries of theory to unlock new pathways for transformative real-world applications. Through exploring the theoretical foundations of modern AI, the team designs robust models and algorithms that power applied innovation.
Our research in the news
See all newsOur Publications
BoostMD – Accelerating MD with MLIP
15 Dec 2024Learning the Language of Protein Structures
15 Dec 2024Bayesian Optimisation for Protein Sequence Design: Back to Basics with Gaussian Process Surrogates
14 Dec 2024SPO: Sequential Policy Optimisation
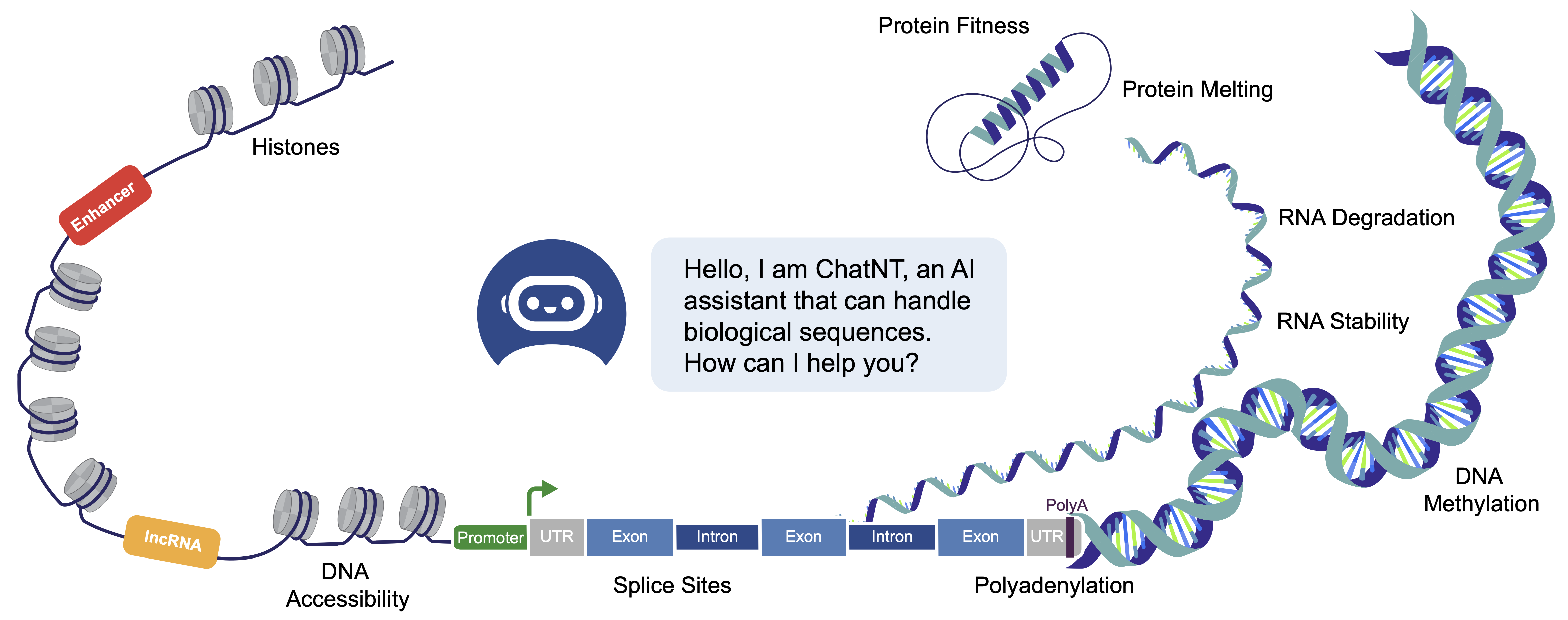
11 Dec 2024Nucleotide Transformer: building and evaluating robust foundation models for human genomics
28 Nov 2024Generative Model for Small Molecules with Latent Space RL Fine-Tuning to Protein Targets
17 Jul 2024Likelihood-based fine-tuning of protein language models for few-shot fitness prediction and design
17 Jul 2024Coordination Failure in Cooperative Offline MARL
17 Jul 2024Machine Learning of Force Fields for Molecular Dynamics Simulations of Proteins at DFT Accuracy
03 May 2024SegmentNT: annotating the genome at single-nucleotide resolution with DNA foundation models
14 Mar 2024How much can change in a year? Revisiting Evaluation in Multi-Agent Reinforcement Learning
13 Mar 2024Preferential Bayesian Optimisation for Protein Design with Fine-Tuned Protein Language Model Ensembles
15 Dec 2023Are we going MAD? Benchmarking Multi-Agent Debate between Language Models for Medical Q&A
15 Dec 2023Optimizing Empty Container Repositioning and Fleet Deployment via Configurable Semi-POMDPs
01 Dec 2023QDax: A Library for Quality-Diversity and Population-based Algorithms with Hardware Acceleration
07 Aug 2023The Quality-Diversity Transformer: Generating Behavior-Conditioned Trajectories with Decision Transformers
17 Jul 2023MAP-Elites with Descriptor-Conditioned Gradients and Archive Distillation into a Single Policy
17 Jul 2023Neuroevolution is a Competitive Alternative to Reinforcement Learning for Skill Discovery
01 May 2023Reinforcement Learning for Branch-and-Bound Optimisation using Retrospective Trajectories
13 Feb 2023Off-the-Grid MARL: Datasets and Baselines for Cooperative Offline Multi-Agent Reinforcement Learning
01 Feb 2023Flow Annealed Importance Sampling Bootstrap
02 Dec 2022Assessing Quality-Diversity Neuro-Evolution Algorithms Performance in Hard Exploration Problems
09 Jul 2022Multi-Objective Quality Diversity Optimization
09 Jul 2022On pseudo-absence generation and machine learning for locust breeding ground prediction in Africa
06 Dec 2021Scaling Properties of Deep Residual Networks
27 May 2021Offline Reinforcement Learning Hands-On
01 Dec 2020Masakhane — Machine Translation For Africa
27 Mar 2020
There are no results that match your search.
Please try different search criteria.